Test Your Knowledge
Cancer Genomics Consortium Quiz Questions
The Cancer Genomics Consortium (CGC) provides a valuable collection of quiz-style questions and answers covering a wide range of topics, from fundamental cytogenetic and genomic principles to advanced applications in cancer and inherited disease diagnostics. Regardless of your role, these questions serve as an engaging way to test your knowledge and reinforce key concepts. By integrating real-world scenarios, the CGC question collection supports ongoing education.
New quiz-style questions are posted on the CGC’s social media channels (X, LinkedIn, Facebook) once a month, offering regular opportunities for learners to engage with fresh content and stay up to date with the latest in clinical laboratory genetics and genomics.
Q33. A young man underwent hereditary cancer testing because of family history. He was found to have a pathogenic variant in promoter region of APC. Which of the following risk is increased in this patient?
A. Attenuated colon polyps
B. Colon cancer
C. Gastric cancer
D. Osteomas
Correct answer: C
Explanation of answer: Promoter region variants are associated with Gastric Adenocarcinoma and Proximal Polyposis of the Stomach (GAPPS). It has 13%-25% lifetime risk for gastric carcinoma.
Q32. In AML with t(8;21), KIT mutations are associated with:
A. Good prognosis
B. Adverse prognosis
C. Standard prognosis
D. Controversial prognosis
Correct answer: B
Explanation of answer: KIT mutations are commonly detected in core binding factor AMLs [t(8;21) and inv(16)]. In t(8;21), KIT mutations are significantly associated with adverse prognosis; however, in inv(16), KIT mutations are not as prognostically significant.
Q31. Mr. Bean has recently been diagnosed of acute myeloid leukemia at 75 years old. His two brothers also had Hodgkin lymphoma and multiple myeloma. Which of the following genes might explain his personal and family history?
A. FANCA
B. TP53
C. MITF
D. DDX41
Correct answer: D
Explanation of answer: DDX41 gene encodes a RNA helicase and germline pathogenic variants are at increased risk to develop hematological malignancies especially AML (PMID: 26712909). Other reported malignancies include CML, Hodgkin lymphoma, non-Hodgkin lymphoma, and multiple myeloma (PMID: 26712909, 31484648)
Q30. What genetic alteration is standard risk in multiple myeloma?:
A. Deletion 13 by karyotype
B. t(11;14)
C. Deletion 17p
D. t(4;14)
Correct answer: B
Explanation of answer: All the above are high/adverse risk in MM except t(11;14) (CCND1::IGH) rearrangements.
Q29. Increased dietary folic acid intake by women in childbearing age has led to ______ % reduction in the occurrence of neural tube defects. :
A. 10-30%
B. 30-50%
C. 50-70%
D. 70-90%
Correct answer: C
Explanation of answer: UK MRC performed a clinical trial in 1991, using 4 mg/day of folic acid and showed approximately 70% reduction in the occurrence of NTDs. The mechanism by which folic acid works is still under investigation, with evidence suggesting its role in nucleic acid synthesis and methylation pathways.
Q28. What is NOT a possible explanation for a deletion of one signal (green or orange) of a break apart probe?:
A. Typical result
B. Regional deletion
C. Deletion causing fusion
D. Unbalanced rearrangement
Correct answer: A
Explanation of answer: Break apart probes are expected to result in a split signal; however, a deletion of either the 5' or 3' signals may in fact be the result of an unbalanced rearrangement (split, then lost the unimportant rearrangement), a deletion of a region that brings either the 5' or 3' end close to a fusion partner on the same chromosome, or just a regional deletion that happens to remove the 5' or 3' signal but not both signals. When you get this result, one must research the pattern to see if it has been reported before and potentially suggest other studies to determine if there is a true fusion (e.g. NGS fusion panels).
Q27. A 16 y.o. female has hypercalcemia and hyperparathyroidism and family history of pituitary gland tumor and hyperparathyroidism in father. What would be likely finding in hereditary cancer panel?:
A. MEN1
B. MEN2
C. MSH3
D. RET
Correct answer: A
Explanation of answer: MEN1 encodes a nuclear scaffold protein, menin, which is involved in regulating gene expression and genomic integrity (PMID:23850066). Pathogenic germline variants in MEN1 are associated with multiple endocrine neoplasia type 1 (MEN1), a autosomal dominant condition features with parathyroid tumor, gastroenteropancreatic neuroendocrine tumor, and anterior pituitary tumor (PMID:22723327, 20301710).
Q26. Which of the following abnormalities are defined as high risk in newly diagnosed patients with multiple myeloma?:
A. 1p32 gain
B. t(4;14)
C. t(11;14)
D. Trisomy 8
Correct answer: B
Explanation of answer: NCCN guidelines v2 2024 (November 1, 2023): del(17p) and/or translocation t(4;14) and/or translocation t(14;16) are considered as high risk in patients with multiple myeloma (MM).
Deletion, and NOT a gain, of 1p32 is among typical findings in MM; t(11;14) is associated with an intermediate risk for survival time; trisomy 8 carries poor prognosis in patients with acute myeloid leukemia but is a rare finding in patients with MM.
Q25. A diffuse astrocytic glioma arising in 65-year old patient is found to be IDH-wild type and H3-wild type, what is a defining genetic feature of glioblastoma, IDH-wild type, CNS WHO grade 4? :
A. Gain of chromosome 10
B. TERT promoter mutation
C. EGFR gene amplification
D. B and C
Correct answer: D
Explanation of answer: Glioblastoma is the most frequent malignant brain tumour in adults, accounting for approximately 15% of all intracranial neoplasms. In children, it accounts for approximately 3% of all CNS tumours. Glioblastoma, IDH-wildtype, is a diffuse, astrocytic glioma that is IDH-wildtype and H3-wildtype and has one or more of the following histological or genetic features: microvascular proliferation, necrosis, TERT promoter mutation, EGFR gene amplification, combined +7/−10 chromosome copy-number changes (CNS WHO grade 4).
Q24. Prader-Willi syndrome result from ______ in a majority of cases.
A. Imprinting defect
B. UPD15
C. Deletion
D. Path seq variant in UBE3A
Correct answer: C
Explanation of answer: Proportion of PWS attributed by mechanism: 15q deletion (60-70%), UP15 complete isodisomy (4-5%), UPD15 segmental isodisomy (17-23%), UPD15 heterodisomy (8-11%), Imprinting center deletion (<0.5%), ICR epimutation (2-4%).
Q23. A 4 year old girl came for genetic testing for malignant neoplasm of peripheral nerves (two tumors next to spine and adrenal gland) and constipation. Which gene will be likely found to be altered?
A. CDH1
B. PHOX2B
C. NF1
D. POLE
Correct answer: B
Explanation of answer: Germline pathogenic changes in PHOX2B are associated with autosomal dominant Congenital Central Hypoventilation Syndrome (CCHS). It was reported to have increased risk for Hirschsprung disease and neuroblastoma (PMID: 15657873, 20301600). Clinical presentation varies due to variable expressivity, reduced penetrance, and somatic mosaicism (PMID: 16888290, 27485184)
Q22. The Pedigree in the image shows which type of inheritance pattern:
A. Autosomal dominant
B. X-linked dominant
C. X-linked recessive
D. Mitochondrial
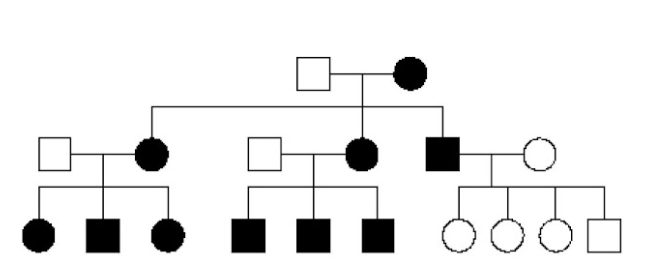
Correct answer: D
Explanation of answer: The pedigree shows a mitochondrial maternal inheritance pattern, where the females in the family pass the disorder to the next generation, but the male in generation II cannot pass it to the next generation.
Q21. OncoScan SNP microarray detected loss of whole arm of chromosome 1p, loss of whole arm of chromosome 19q and sequencing detected an IDH1 p.Arg132His mutation in a brain sample from a patient with seizures and headache. These findings confirm the diagnosis of:
A. Astrocytoma
B. Oligodendroglioma
C. Diffuse midline glioma
D. Glioblastoma
Correct answer: B
Explanation of answer: Oligodendrogliomas are molecularly defined by IDH1 or IDH2 mutations and 1p/19q codeletion, and only complete whole arm losses of both chromosome arms are diagnostic for oligodendroglioma (Louis DN, et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol. 2021 Aug 2;23(8):1231-1251. PMID: 34185076)
Q20. All tumor types below have BRAF V600E, except:
A. Hairy cell leukemia
B. Colon adenocarcinoma
C. Melanoma
D. Lymphoplasmacytic lymphoma
Correct answer: D
Explanation of answer: All the others (HCL, Melanoma, CRC) recurrently demonstrate BRAF V600E. Lymphoplasmic lymphoma is associated with MYD88 mutations, specifically L265P.
Q19. What cancer type would you perform methylation analysis on?
A. Brain
B. Endometrial
C. Lung
D. Sarcoma
Correct answer: A
Explanation of answer: Methylation analysis on brain tumors has proven to be an accurate diagnostic tool.
Q18. The most common mechanism of inherited cancer predisposition is:
A. Oncogenic alteration
B. Polymorphism
C. Path variant with 2nd hit
D. SNP with second hit
Correct answer: C
Explanation of answer: The most common mechanism for inherited cancer predisposition is a pathogenic germline variant and then an acquired second hit on the wild type allele.
Q17. A patient phenotypically is presenting with Prader-Willi. What testing is diagnostic for PWS?
A. Bisulfite-treatment PCR
B. Karyotype
C. Chromosomal Microarray
D. FISH
Correct answer: A
Explanation of answer: While FISH, CMA, and sometimes karyotype (depending on resolution) can indicate a deletion in the region of PWS, only methylation sensitive PCR can identify which parental allele that the deletion resides on
Q16. What defines the threshold for quantitative real-time PCR?
A. Amount after 10 cycles
B. #of cycles for log-linear
C. Slope of amp curve
D. #of cycles for plateau
Correct answer: B
Explanation of answer: The cycle-threshold is defined as the fractional cycle number in the log-linear region of PCR amplification in which the reaction reaches fixed amounts of amplicon DNA.
Q15. Which karyotype if found in a 32-year-old woman with a normal phenotype, would explain her history of recurrent pregnancy loss?
A. 45,X
B. 46,XX
C. 46,XX,+13,der(13;14)
D. 46,XX,t(11;13)
Correct answer: D
Explanation of answer: Balanced translocations are a common finding in normal phenotype parents with recurrent pregnancy loss.
Q14. What is the PRIMARY reason that the cytogenetics lab needs to know the possible diagnosis when processing a newly received bone marrow sample?
A. CAP requirement
B. Cell culture conditions
C. Know amount needed
D. Reimbursement
Correct answer: B
Explanation of answer: Knowing the reason for referral is important for cell culture selection.
Q13. A 5 yr old being evaluated for ALL. The ped onc orders FISH: nuc ish(ABL,BCR)x2,(KMT2Ax3),(ETV6x3,RUNX1x4) [166/200]. What does it mean?
A. Likely mosaic for triploid line
B. Positive for ETV6::RUNX1
C. Suggestive of hyperdiploidy
D. Evidence of iAMP21 B-ALL
Correct answer: C
Explanation of answer: When multiple probes demonstrate copy gains, the most likely answer is a hyperdiploid karyotype; however, it is important to demonstrate this subtype by either multiple centromere probes (FISH) or by karyotype.
Q12. A 70 yo female with suspected MDS has the result: 46,XX,del(5)(q13q33)[4]/47,XX,+8[4]/46,XX[12]. What does this mean?
A. 40% had del chr 5, extra chr 8
B. There were two unrelated abnormal clones
C. There were two related abnormal clones
D. 40% had dels of chrs 5 & 8
Correct answer: B
Explanation of answer: This is a question about how to read the ISCN properly. Because there is no indication of acknowledgment of a stemline in the second set of cells, this indicates that there are two separate abnormal populations of cells comprising a total of 40% of the cells. The remaining 12 (60%) are normal. While one may expect to see the clones as related in disease, it is possible the two lines either appeared separately or they may represent subclones in the main disease line with another genetic driver (e.g. SF3B1 as the driver with one subclone with del(5q) and the other subclone +8). Using just the karyotype, one cannot differentiate between these two possibilities.
Q11. What tumor type is likely to generate this karyotype?
A. Osteosarcoma
B. Alveolar rhabdomyosarcoma
C. Synovial Sarcoma
D. Atypical lipomatous tumor
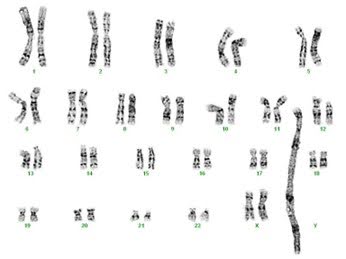
Correct answer: B
Explanation of answer: Amplification of MDM2, CDK4 can be useful to distinguish Atypical lipomatous tumor/well-differentiated liposarcoma and Dedifferentiated liposarcoma from benign tumors. These amplifications are generally detected as supernumerary rings and giant marker chromosomes on karyotype.
Q10. Paired tumor/normal NGS was performed on a bone marrow specimen from a pediatric patient with cytogenetically normal acute myeloid leukemia. A CEBPA mutation was identified, c.175G>T p.(Glu59Ter). This finding suggests that the patient has:
A. A favorable prognosis
B. A germline CEBPA variant
C. An undetected 3' mutation
D. A high p30:p42 ratio
Correct answer: D
CEBPA-mutated acute myeloid leukemia (AML) is its own diagnostic category within the WHO Classification of Haematolymphoid Tumors. Current literature supports the presence of CEBPA biallelic mutations (i.e., biCEBPA) or single mutations located within the 3' basic leucine zipper (bZIP) region of the gene (i.e., smbZIPCEBPA) as favorable, whereas single mutations found outside of this bZIP region do not have a more favorable prognosis (as compared with CEBPA-wildtype AML). Thus, answer A would not be correct. N-terminal CEBPA mutations have been shown to occur in the germline of individuals with AML at a higher frequency than variants located within other regions of the gene. Although in a tumor-only test setting the location of this variant in the N-terminus of CEBPA would support further investigation into the origin of this variant, the paired tumor/normal design of the NGS assay confirms that this N-terminal variant is somatic. Thus, answer B would not be correct. Although biallelic CEBPA mutations are commonly identified in CEBPA-mutated AML, single mutations are not uncommon. Thus, although technically possible, answer C is not the best answer. The CEBPA protein exists as two primary isoforms, a full-length isoform (p42) and a shorter isoform (p30). Due to the fact that CEBPA has two translation initiation sites, the presence of a truncating N-terminal variant often results in a decrease of the p42 isoform and increase in the p30 isoform. This leads to an increased p30:p42 ratio which drives an immature cell state. Thus, D is the correct answer.
Q9. A young patient presented with a thigh mass. Histologic examination showed a small round blue cell tumor. FISH studies with the EWSR1 break-apart probe were performed. Based on the result of the FISH studies, select the correct answer:
A. This is a normal result
B. Rearrangement of EWSR1
C. Rhabdomyosarcoma
D. Synovial sarcoma
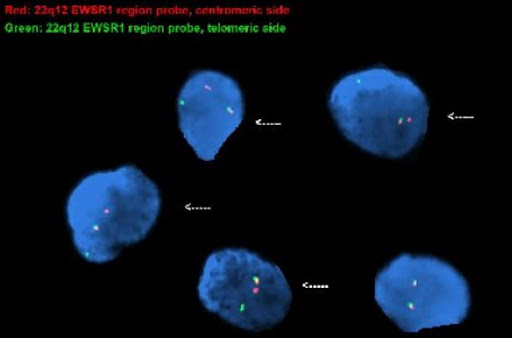
Correct answer: B
Explanation of answer: This is an abnormal signal pattern consisting of one intact (fused) signal, one 5’ (centromeric) signal, and one 3’ (telomeric) signal indicating a rearrangement of 22q12 (EWSR1) region; consistent with the diagnosis of Ewing’s sarcoma. PAX3::FOXO1 or PAX7::FOXO1 translocations are present in alveolar rhabdomyosarcoma. Spindle cell / sclerosing rhabdomyosarcoma may harbor MYOD1 mutations or VGLL2::NCOA2 rearrangements. Synovial sarcoma is characterized by a specific SS18::SSX fusion gene.
Q8. A 4-year-old boy had bilateral lung cysts and cystic nephroma. A splicing variant was detected in a hereditary cancer NGS panel and tumor biopsy showed another frameshift variant in the same gene at 15%. What function is the gene involved?
A. DNA mismatch repair
B. Homologous recombination
C. RNA splicing
D. Gene silencing
Correct answer: D
The clinical presentation suggests the causative gene is DICER1 which encodes a protein that facilitates microRNA production. microRNA is an important mechanism that regulates gene expression by silencing mRNA at post-transcription level.
Q7. New B-lymphoblastic leukemia case: Based on genetics what result(s) has a very favorable prognosis?
A. t(4;11)(q21;q23)
B. t(12;21)(p13.2;q22.1)
C. Hypodiploid karyotype
D. t(9;22)(q34.1;q11.2)
Correct answer: B
Out of these options, for B-ALL t(12;21) is the only favorable prognosis. Hyperdiploid karyotype is also a favorable prognosis.
Q6. What FISH panel would you order for myeloid neoplasms?
A. Chr. 5, 7, 8, 17, 20
B. Chr. 11, 12, 13, 17
C. Chr. 5, 11, 13, 17, 20
D. Chr. 1, 13, 14, 17
Correct answer: A
Alterations commonly seen in myeloid malignancies include deletion 5q, 7q, monosomy 5, 7, trisomy 8, deletion 20q and deletion 17p/monosomy 17.
Q5. During a surgical pathology rotation, you are asked to send a sample on a patient who might have a soft tissue tumor for cytogenetic G-band analysis. Which of these samples would you send to the lab?
A. Blood
B.Fresh tissue
C.Frozen tissue
D.Formalin fixed tissue
Correct answer: B
For a solid tumor, such as soft tissue tumor, the best tissue for cytogenetic testing by karyotype is the fresh tumor tissue. The FFPE is fixed and therefore cannot be cultured. The frozen will most likely not grow as the cells are dormant/dead. The blood will not have tumor cells that can be cultured.
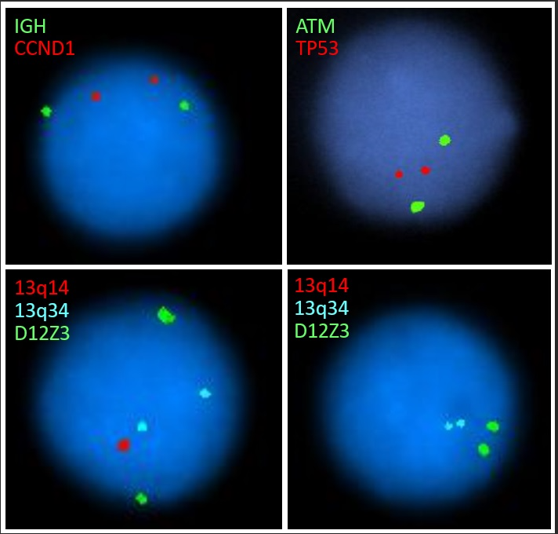
Q4. The following CLL FISH panel result is from a 77 year old man referred due to an elevated white blood cell count. Based on the representative images, which statement is correct?
A.This result is consistent with mantle cell lymphoma
B.This result is associated with a good prognosis in CLL
C.This result is not specific for CLL
D.This result is associated with the most adverse prognosis in CLL
Correct answer: B
This patient has both heterozygous and homozygous deletion of 13q14. This is a common finding in CLL. Patients with a 13q deletion as the sole abnormality detected by FISH generally have the longest survival time. Prognosis for patients with a homozygous 13q deletion is not significantly different than for a heterozygous 13q deletion (PMID: 22139735). Some newer data suggests that larger deletions may have less favorable outcomes (PMID: 34522485).
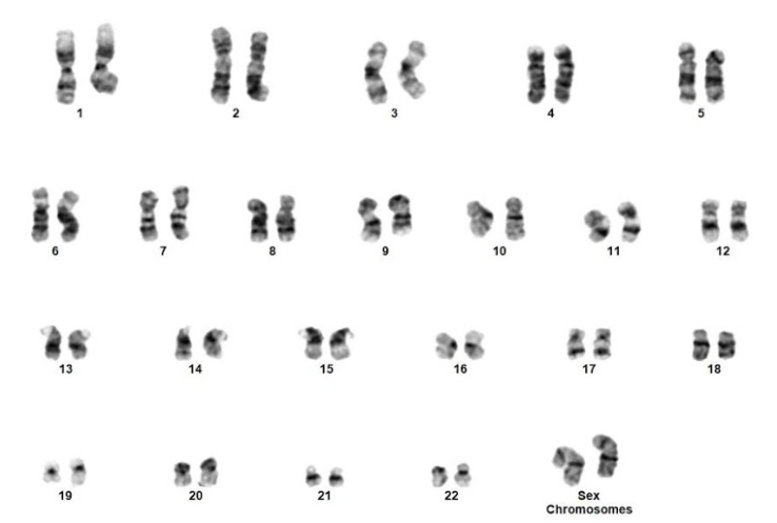
Q3. A female with a history of AML post transplant with a male donor has presented with the following karyotype. Which of the following is true based on the karyotypic abnormality?
A. Associated with a good prognosis for the patient
B. Auer rods are commonly present
C. Basophilia is commonly present
D. Monosomy 7 is a common secondary finding
Correct answer: D
The inv(3)(q21q26) is often associated with a fusion between RPN1 and MECOM (MDS1 AND EVI1 COMPLEX LOCUS). It has been described in AML and MDS. Monosomy 7 is a common secondary abnormality. It is associated with a negative impact on prognosis.
Q2. According to the American College of Medical Genetics Laboratory Technical Standards and Guidelines, how long should images from neoplastic FISH cases be retained?
A. Two weeks after the report has been signed
B. Three years
C. Ten years
D. Twenty years
Correct Answer: C
In general, "critical records" for cytogenetic testing are kept for 20 years. However, there is variability depending on the technology utilized and reason for referral.
Current guidelines specify the following retention times:
- Processed patient specimens and/or cell pellets – 2 weeks after report has been signed
- Stained slides (e.g., G-banding) – 3 years
- Neoplastic FISH Images – 10 years
- Images (chromosome analysis/non-neoplastic FISH) – 20 years
- Array – recommended that primary data be stored for at least 2 years and the analysis file + final clinical report be kept for “as long as possible”
(Shao, et al., 2021)
Q1. NGS has been widely used to identify germline variants for diagnosing hereditary cancer syndromes. However, it may also detect mosaic variants that may be acquired due to clonal hematopoiesis of intermediate potential (CHIP). Which of the following cancer genes is most likely to be involved in CHIP?
A. TP53
B. DICER1
C. PTEN
D. BRCA2
Correct answer: A
CHIP is associated with aging in hematopoietic stem and progenitor cells and the most commonly variants have been seen in DNMT3A, TET2, ASXL1, JAK2 and TP53. TP53 is one of the most commonly tested gene in hereditary cancer panel, therefore, skewed allele frequency in TP53 may indicate an event of CHIP, which needs to be differentiated from Li-Fraumeni syndrome. PMID: 31827082